
MANGEM: A web app for multimodal analysis of neuronal gene expression, electrophysiology, and morphology
Recently, it has become possible to obtain multiple types of data (modalities) from individual neurons, like how genes are used (gene expression), how a neuron responds to electrical signals (electrophysiology), and what it looks like (morphology). These datasets can be used to group similar neurons together and learn their functions, but the complexity of the data can make this process difficult...

Joint Variational Autoencoders for Multimodal Imputation and Embedding
Single-cell multimodal datasets have measured various characteristics of individual cells, enabling a deep understanding of cellular and molecular mechanisms. However, multimodal data generation remains costly and challenging, and missing modalities happen frequently. Recently, machine learning approaches have been developed for data imputation but typically require fully matched multimodalities...
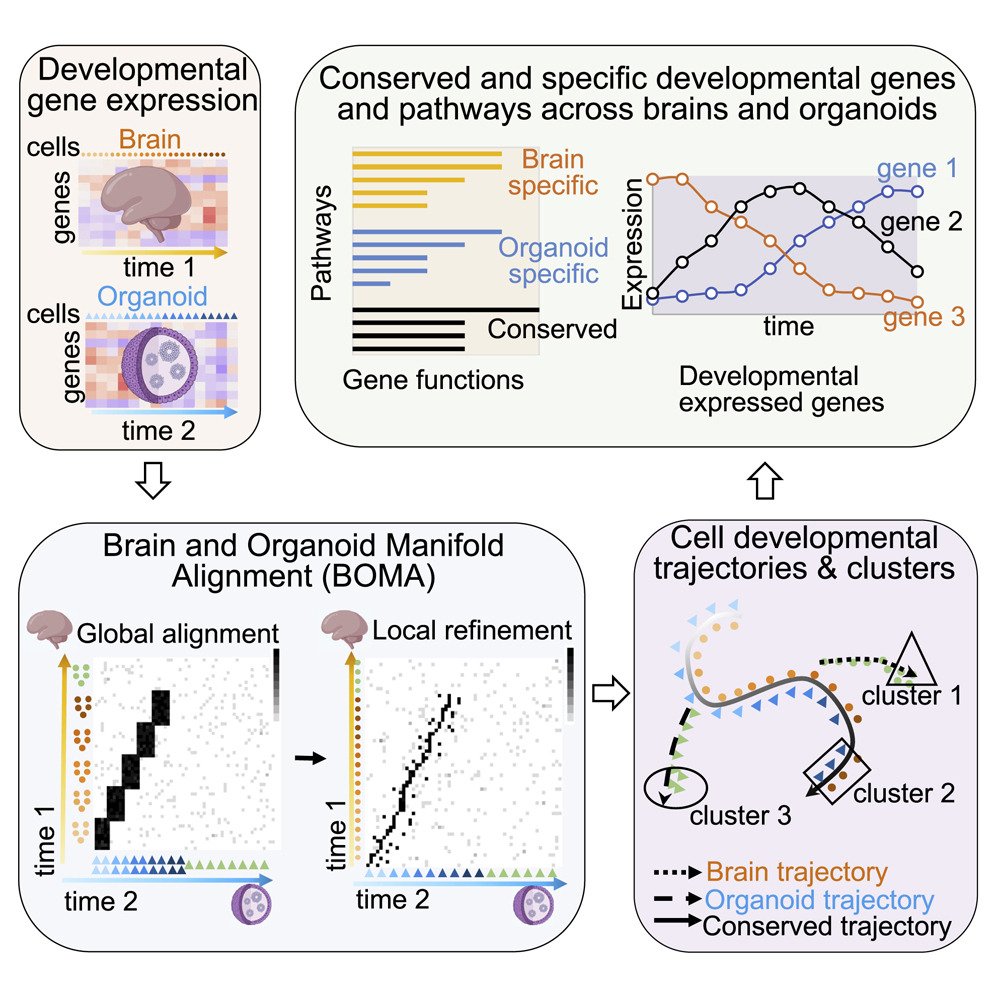
BOMA, a machine-learning framework for comparative gene expression analysis across brains and organoids
Organoids have become valuable models for understanding cellular and molecular mechanisms in human development, including development of brains. However, whether developmental gene expression programs are preserved between human organoids and brains, especially in specific cell types, remains unclear. Importantly, there is a lack of effective computational approaches for comparative data analyses...