
COSIME: Cooperative multi-view integration and Scalable and Interpretable Model Explainer
Single-omics approaches often provide a limited view of complex biological systems, whereas multiomics integration offers a more comprehensive understanding by combining diverse data views. However, integrating heterogeneous data types and interpreting the intricate relationships between biological features—both within and across different data views—remains a bottleneck. To address these challenges...

Network-based drug repurposing for psychiatric disorders using single-cell genomics
Neuropsychiatric disorders lack effective treatments due to a limited understanding of underlying cellular and molecular mechanisms. To address this, we integrated population-scale single-cell genomics data and analyzed cell-type-level gene regulatory networks across schizophrenia, bipolar disorder, and autism (23 cell classes/subclasses). Our analysis revealed potential druggable transcription...

Personalized Single-cell Transcriptomics Reveals Molecular Diversity in Alzheimer’s Disease
Precision medicine for brain diseases faces many challenges, including understanding the heterogeneity of disease phenotypes. Such heterogeneity can be attributed to the variations in cellular and molecular mechanisms across individuals. However, personalized mechanisms remain elusive, especially at the single-cell level. To address this, the PsychAD project generated population-level single-nucleus...

NeuroTD: A Time-Frequency Based Multimodal Learning Approach to Analyze Time Delays in Neural Activities
Studying temporal features of neural activities is crucial for understanding the functions of neurons as well as underlying neural circuits. To this end, recent researches employ emerging techniques including calcium imaging, Neuropixels, depth electrodes, and Patch-seq to generate multimodal time-series data that depict the activities of single neurons, groups of neurons, and behaviors. However...

MANGEM: A web app for multimodal analysis of neuronal gene expression, electrophysiology, and morphology
Recently, it has become possible to obtain multiple types of data (modalities) from individual neurons, like how genes are used (gene expression), how a neuron responds to electrical signals (electrophysiology), and what it looks like (morphology). These datasets can be used to group similar neurons together and learn their functions, but the complexity of the data can make this process difficult...

Joint Variational Autoencoders for Multimodal Imputation and Embedding
Single-cell multimodal datasets have measured various characteristics of individual cells, enabling a deep understanding of cellular and molecular mechanisms. However, multimodal data generation remains costly and challenging, and missing modalities happen frequently. Recently, machine learning approaches have been developed for data imputation but typically require fully matched multimodalities...
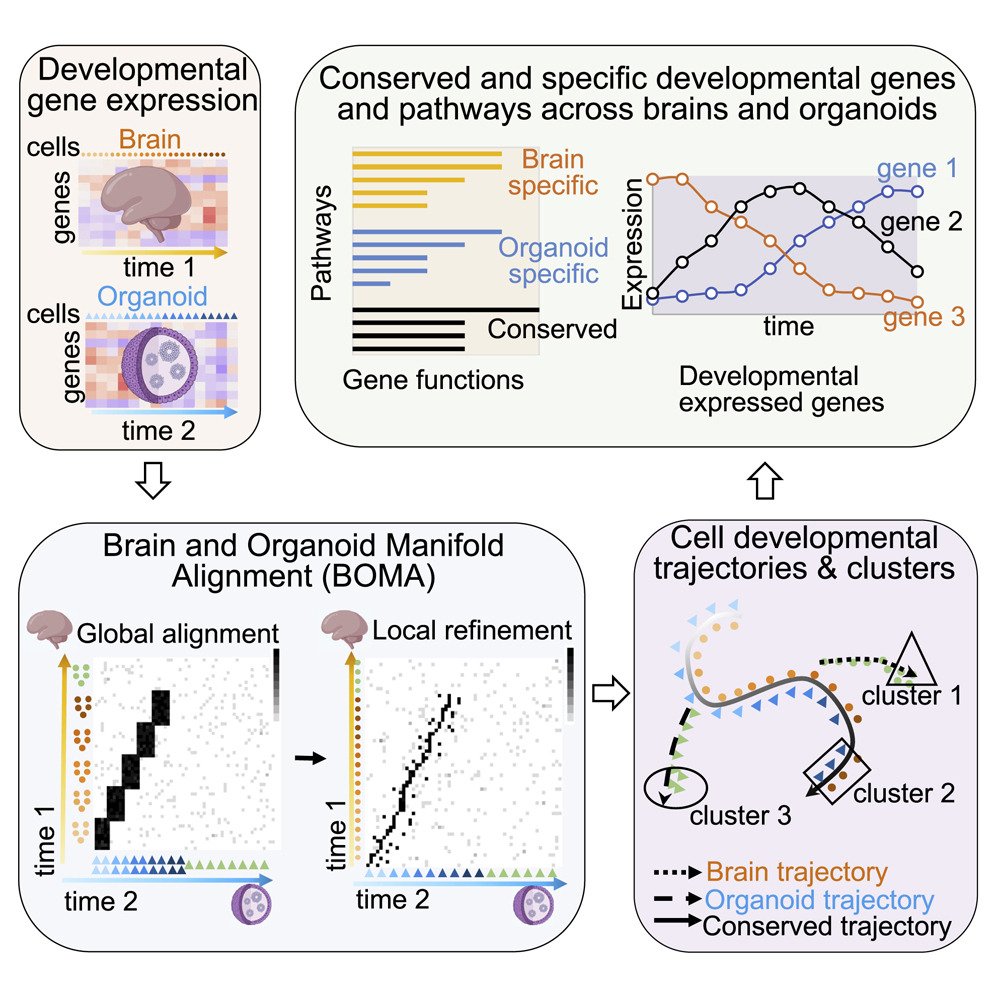
BOMA, a machine-learning framework for comparative gene expression analysis across brains and organoids
Organoids have become valuable models for understanding cellular and molecular mechanisms in human development, including development of brains. However, whether developmental gene expression programs are preserved between human organoids and brains, especially in specific cell types, remains unclear. Importantly, there is a lack of effective computational approaches for comparative data analyses...